Contents
- Overview
- After a sequencing job is complete, relevant contacts (both researcher and Lab contact, if available) will receive an email with links to the fastq data for your sample(s). There you will find buttons to download the data to your computer and to upload the data to Galaxy. The data will remain in HTSEQ indefinitely and you can return to it anytime.
- Individual sample, assay and dataset pages all contain buttons to upload and download your data. In addition, assay pages contain links allowing you to download a single file at a time.
- A set of samples, assays or datasets resulting from a Search or the menu options "My Samples", "My Assays", and "Viewable Datasets" contain buttons to download data for all the checked items.
- Batch Data Retrieval Page
- Downloading Data to Your Computer
- Multiple Files
- Individual Files
- Uploading Data to Galaxy
- From HTSEQ
- From Galaxy
- Log into Galaxy.
- Click the "Princeton HTSEQ Data Import" link beneath the left-hand "Tools->Get Data" menu
- Find the targeted sample, assay, or dataset record within HTSEQ
- Within the "Raw Data" table, each archived file has a dedicated row; click the button for the file of interest.
- Sharing Your Data with a Collaborator
The HTSEQ data model consists of samples, assays, and derived datasets. Samples are sequenced with assays as their output. Assays contain all the raw fastq data (both passed and failed reads) from the sequence run as well as fastqc files that provide basic QC analysis. If a sample is sequenced on more than one lane of a flowcell, individual assays will be generated and their raw data combined into a merged dataset. If your , then the data is split apart into a demultiplexed dataset with fastq files for each sample. (For 10X samples, two additional datasets can be created by processing your data through the Cell Ranger pipelines:Cell Ranger Count and Cell Ranger Aggregate.)
Your data is accessible in HTSEQ through a variety of avenues:
The Batch Data Retrieval page provides fastq files for only the passed, non-indexed reads for assays and datasets. Individual fastq files for indexed and/or failed reads are available on the Assay page's raw data table.
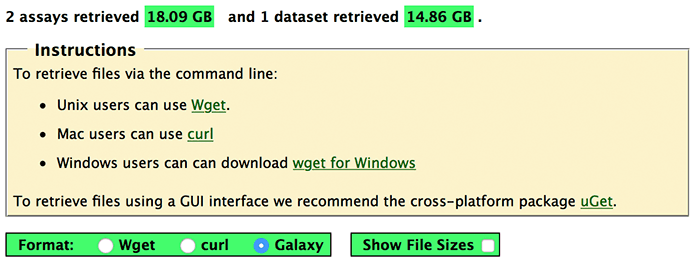
The top portion of the Batch Data Retrieval page lists the number of assays and datasets retrieved along with instructions and a set of radio buttons that allow you to specify the format of the URLs.
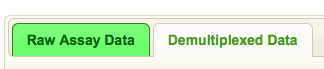
Data is display in tabs. Raw assay data is displayed in one tab, and derived
data is displayed in additional tab(s). For example, if your sample(s) are
barcoded you will see two tabs, one for the raw assay data and one for the demultiplexed data:
If you are downloading data to a Mac we provide URLs for the curl program which is built into OS X. There is one URL per file which you copy and paste into a terminal window on your Mac.
Wget is an alternative option for retrieving files, available from here. Unlike curl, wget can download an entire set of files with a single URL, so there is less copy and pasting involved.
If you prefer a GUI interface then we recommend the cross-platform uGet, an open-source download manager.
The assay page contains links to all the raw data files as well as fastqc statistics files, allowing you to download a single file at a time.
In addition to fastq files, you can download the barcode file and we also provide a md5sum file so that you can check the integrity of the download. All files available for download or upload to Galaxy are represented as formatted URLs.
You can upload your data to Galaxy in two ways. The simplest way is to select the files you want to upload and then click on the "Upload Selected to Galaxy" button (please log into Galaxy first). This loads the files into your Galaxy History.
A second method exists for demultiplexed data which allows you to load your data
into a Galaxy Rule-based Collection. Copy the table that appears at the
bottom of the demultiplexed tab. Log into Galaxy and click the
 button at the top left. A pop-up box appears with tabs.
Select the "Rule-based" tab, paste the table and press "Build". Additional instructions can be found in
this tutorial.
button at the top left. A pop-up box appears with tabs.
Select the "Rule-based" tab, paste the table and press "Build". Additional instructions can be found in
this tutorial.
To share your data with a collaborator, first ascertain how they want their data delivered: via curl on a Mac, wget commands on unix, or plain urls, then select the appropriate "Format" button (curl, wget or Galaxy, respectively). Copy the links as described above and email them to your collaborator. Your collaborators do not have to have accounts on HTSEQ in order to retrieve the data in this manner. Note that the links will be active for only one month.
